April 15, 2019
Exploiting the same force that causes raindrops to cling to windows, Cristian Soitu creates robust fluid walls and microfluidic arrangements using a simple, quick and inexpensive method to create multi-chamber grids and complex circuits.
“The researcher can move from imagining a fluidic circuit to actually building and using it in less than an hour,” says Cristian Soitu, M.Eng., a Ph.D. candidate at University of Oxford, Oxford, United Kingdom (U.K.). Soitu describes a novel fluid-shaping technology built by his team at Oxford that gives biologists the benefit of microfluidic methods while employing familiar materials – cell media and Petri dishes.
"In the regular-sized world, we use solid structures to hold liquids because gravity is a powerful force working against keeping liquid in place," says Soitu. “Solid walls are also used in conventional microfluidics. The microworld, however, offers an additional option. When things get very small, gravity becomes negligible and interfacial forces dominate allowing us to form structures using liquid walls.”
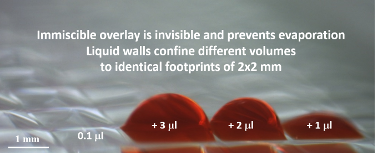
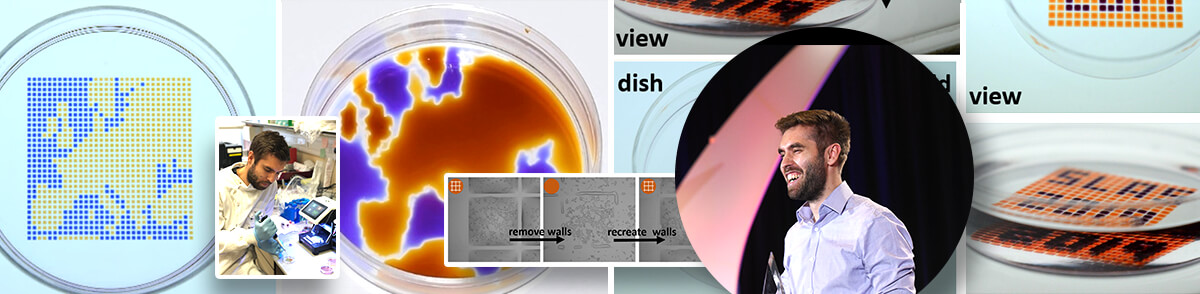
To achieve this, Soitu first covers the surface of a Petri dish with a thin layer of cell medium and overlays it with an immiscible fluorocarbon (FC40), an oil that prevents evaporation. He then creates liquid walls by applying a Teflon stylus to reshape the two layers into an array of individual chambers. The method does not require a dedicated facility or specialized equipment. Instead researchers may use a syringe pump and an automated positioning system to drive the stylus to build fluid walls accurately and reproducibly, scaling up the chamber density as needed. “This contrasts with conventional polydimethylsiloxane (PDMS) circuits where you have to place an order and wait a few weeks until they are produced,” he comments.
The technology was initially conceived by Edmond Walsh, Ph.D., (Oxford Thermofluids Institute, Department of Engineering Science), Alex Feuerborn, Ph.D. (Iota Sciences Ltd.) and Peter Cook, D.Phil., (Oxford Sir William Dunn School of Pathology), after forming an interdisciplinary collaboration to review and change the workflows that cell biologists have used for decades. Soitu joined this team as a D. Phil student (supervised by Prof. Walsh and Alfonso Castrejón-Pita, Ph.D. of Engineering Science, University of Oxford) and was the first to explore the application space, and basic physics of the technology. These were the basis for his award-winning presentation.
Soitu, whose work is funded by an Oxford spin-out company, iota Sciences Ltd., a participant in SLAS2018 Innovation AveNEW, identified some of the obvious shortcomings of existing workflows, which included waste generation, frequent use of large volumes resulting in expensive assays and issues with poor optical properties caused by solid walls in well plates.
“In light of all these factors, we wanted to keep materials involved to a minimum. We did not want any harsh chemicals involved, or any chemical surface treatment,” says Soitu. “The resulting technology essentially empowers researchers to access microscale investigations, and thereby enables the acceleration of existing assays and the development of new ones.” He adds that researchers can anticipate considerable savings in consumables, as well.

To date, this transformative approach to manipulating small volumes has successfully miniaturized many workflows used in cell biology that should offer wide appeal to the biomedical community. “There are so many ways to apply the technology,” continues Soitu, adding that the cross-disciplinary nature of this work provides an excellent foundation for translation to academic and industrial biomedical labs. “At the moment, I am developing a novel way of making these fluidic structures that gives us even more accuracy, opening up many applications that we hadn’t yet explored. I do this because it’s exciting, but at a level beyond the individual, I believe such technologies are critically important to address major societal challenges, such as drug discovery.”
The novel concept earned Soitu the SLAS Innovation Award at the SLAS2019 International Conference and Exhibition (Washington, DC, USA) for his exceptional podium presentation, “Microfluidic Chambers Using Fluid Walls for Cell Biology.” Soitu accepted a $10,000 cash prize as prize as part of this prestigious award.
Soitu demonstrates the versatility of his arrays of chambers (GRIDs) - analogs of a common platform in cell biology, the microtiter plate. His technology captures many of the functions required for cell culture and downstream analysis, including feeding, replating, cloning, cryopreservation, lysis plus RT-PCR, transfection plus genome editing and fixation plus immunolabeling (when fluid walls are reconfigured during use). The technology supports the growth and normal response to stimuli of mammalian cells and the development of worm eggs into adults.
Soitu anticipates that GRIDs will have wide appeal to those who want to obtain higher throughput while maintaining submicroliter volumes. “Despite improvements in life sciences' ability to manipulate ever smaller volumes, most workflows in cell biology still use many microliter volumes,” he explains. “Microplates with more than 1,536 wells that have working volumes down to about 3 microliters are rarely used, largely because of problems introduced by evaporation and air bubbles. It’s the same issue with conventional microfluidic devices. The consequence is that hardly anyone uses nanoliter volumes.”
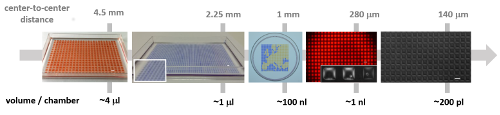
To add uniform volumes quickly and easily, the team investigated methods that did not require washing or replacing of pipette tips. What they discovered was that without solid walls, there is no need to raise, lower or wash the delivering pipette tip or to start and stop the pump. Instead, one tip can scan through FC40 at a constant speed and height, ejecting liquid continuously to feed chambers through transient liquid bridges. Delivery to many chambers occurs speedily without carryover because the walls of FC40, which always remain in contact with the tip, spontaneously wash the tip and self-heal when the tip is removed. Such scanning can deliver 70 nL to each of 1,024 chambers in 90 seconds.
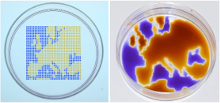
After testing many potential options, Soitu chose FC40, the bio-friendly carrier fluid of choice in droplet-based microfluidics, because it is one of the most inert liquids known and carries vital gases, such as O2 and CO2, so effectively that it has been tested as a blood substitute. Soitu reports that the method works equally well with silicone and hydrocarbon oils, and with glass and polystyrene coated surfaces – a critically important aspect for stem-cell activities. The water-immiscible phase effectively isolates each chamber in the dish preventing contamination and communication between adjacent chambers. For example, when Soitu manually pipetted E. coli into every second chamber in an 8 × 8 cm grid, the bacteria grew only in inoculated drops while the rest remained sterile. “The grids also withstand violent agitation,” he adds.
“Another important aspect of this technology is the ability to reconfigure structures and the environment during an assay without disturbing or harming the cell,” Soitu continues. “We are able to repeatedly remove and recreate fluid walls. Essentially, we are transforming a multi-well plate into a single-well one, and then reverting back to the initial multi-well format. This level of flexibility is unprecedented to our knowledge.”
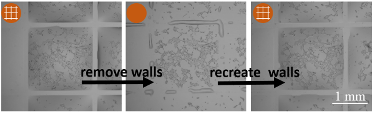
The simple technology provides biologists with a user-friendly microfluidics method that offers excellent physical and optical access to the sample, characteristics that will be particularly valuable, Soitu believes, during cell cloning. “When cells are cloned commercially in the wells of a microplate, they almost invariably grow up against the solid walls at the edge of each well. This edge factor obstructs views, making it impossible to be certain that only one cell is present per well. As a consequence many people perform a second round of cloning,” Soitu explains. “We don’t have edge effects because the fluid walls are completely transparent. Users can be confident that only one cell per chamber is present.” In addition, growing clones can be identified earlier and picked sooner, he adds.
“Working on anything biological was not on my radar during my undergraduate and graduate studies,” says Soitu, who focused on computational methods for fluid flow relations during his studies at the University of Aberdeen (Aberdeen, Scotland, U.K.), where he graduated with a degree in mechanical engineering in 2016. “I had no possible applications in mind before my Ph.D. program.”
He knew that his interest in fluid dynamics lay at the core of most biological processes, and this propelled him to apply for Ph.D. programs with a biomedical focus. After being accepted into the Ph.D. program at the University of Oxford and joining the early-stage, multidisciplinary research in the labs of Walsh and Cook in the fall 2016, Soitu launched into developing his microfluidic platform.
In 2018, after completing much of the work that went into his technology, Soitu had the eye-opening opportunity to attend SLAS2018 in San Diego, CA, USA. “SLAS is definitely one of the best conferences I have attended,” he comments. “The Society is unparalleled in creating a fruitful mixture of academia and industry. Especially for a Ph.D. student like myself, it is incredibly beneficial to be exposed to this intersection of science, engineering and technology. One gets to explore all of these worlds over the course of three days.”
Soitu feels that conference connections are about engaging interests. “I am not a biologist. Sometimes I need to get out there and try to find problems to solve and see what others are doing. It definitely had an impact on my research over the last year,” he says.
In particular, he enjoyed attending the academic tracks that were not that directly related to his project. “I saw a lot of interest in imaging C. elegans, for example,” Soitu says. “I learned more about a microfluidic device that was designed to trap, confine and image the worms. That presentation led me to think about how my system would be ideal for imaging because the method makes it easier to confine the animals and track the progress of the experiment and the development of the worms at the same time.”

This influx of knowledge also influenced his decision to submit his work for the SLAS2019 Innovation Award. “To have one’s work recognized – especially as a graduate student – has tremendous positive impact in advancing my work and this technology,” says Soitu, who is equally excited to be a part of the SLAS2020 Innovation Award jury – a privilege that comes along with winning the prize. “The word is getting around about our interesting technology. People are keen to establish collaborations,” he comments.
With so many avenues to explore, “interactions, such as those I experienced at the SLAS International Conference and Exhibition, are so important for us,” Soitu comments. “People come to us and say, ‘it would be great if you could do this…’ Connecting at the conference helps us identify workflows that we are not aware of that may be useful to many other users. All it takes is a researcher to go through one of our papers, an article or a presentation and apply it to what they are doing. This is what we hope will evolve from sharing our discovery.”
Soitu, who got accepted as a podium presenter for the 2019 SLAS Europe Conference and Exhibition, will give an up-to-date presentation on his work at the event to be held June 26-28 in Barcelona, Spain. “I am constantly working on applications and workflows. I have a few things that I did not include in my Washington, DC, presentation that I hope to include in the presentation for Barcelona.”
Learn More about the SLAS Innovation Award and 2019 Finalists!
From SLAS Technology: Next-Generation Compound Delivery Platforms to Support Miniaturized Biology
Learn More about Microfluidics with Fluid Walls
Microfluidic Chambers Using Fluid Walls for Cell Biology
An SLAS Discovery Review Article: Three-Dimensional Cell Cultures in Drug Discovery and Development
Read about Advances in Micro?uidics and Lab-on-a-Chip Technologies